|
|
|
|
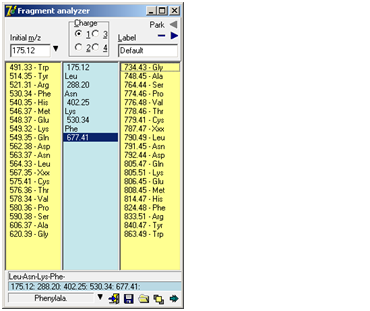
I Download Fragment analyzer. f you try to manually de-novo interpret an ms/ms spectrum of a peptide, you will usually be in for a lot of typing on your calculator. Even if your spectrum software is able to show mass differences on the screen, there can be a lot of mouse movements and calculations. The Fragment Analyzer is a tool, which will help you manually calculate (assign) a series of fragment ions in a simple and flexible way. The ‘Fragment Analyzer’ is a separate program (‘Fragment.exe’) called from GPMAW. If the menu item (Utilities | Fragmen analyzer) and the toolbar icon is grayed and inaccessible, GPMAW has been unable to locate the file. Please copy the file ‘Fragment.exe’ to the \gpmaw\bin\ directory and restart GPMAW: Working with the Fragment analyzer is quick and easy:
As you select a new mass value, the selection in the center box automatically jumps to that selection, going up or down. If you want to cancel part of a series, just select the last entry you want to keep, and then click on the new next entry (up or down) – the rest of the series in the relevant direction will be deleted. The bottom part of the dialog box shows the fragment series, the m/z series, the full name of the currently selected residue and buttons for ‘Options menu’, ‘Exit’, ‘Save fragment’, ‘Load fragment’ and ‘Stay on top’. The ‘Options menu’ is also available as pop-up menu (right-click) and contains the following commands:
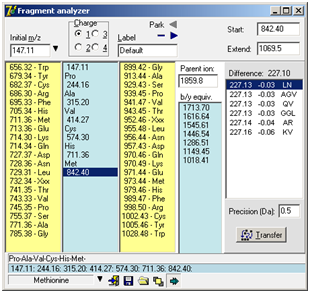
The label is used as filename/title when saving the fragment to disk and when ‘Parking’ the fragment. Park: When trying to interpret a complex ms/ms spectrum, you often get partial sequences in different parts of the spectrum. By using the ‘parking’ area, you can save the list temporarily by pressing the little right-arrow in the top corner of the dialog. This will store the present fragment in a list and make the left-arrow active. Pres the left-arrow to open the list and retrieve a previously stored fragment. Click on the minus ‘-‘ button to clear the fragment list. Extended display mode Clicking on the bottom right button ‘Extended display mode’ will enlarge the ‘Fragment Analyzer’ window to show the b/y equivalent column and the ‘Extend sequence’ list box. b/y ion equivalent: An additional column with an input field labeled ‘Parent ion’ at the top is displayed to the right of the ‘up mass residues’. Upon entering the parent ion mass of the peptide in question, the b/y ion equivalent series of mass values relative to the mass values in the center column will be displayed. Whenever a change occurs in the mass fragment column or the parent ion field, the list will be updated. The b/y ion equivalent is the series that is complementary to the one entered in the center column. I.e. if the series entered in the center column originates from the y ion series, the list presented here is the corresponding b ion series. Seeing both the y and the b ion in a spectrum gives more confidence to the assignment.
The ‘Start value’ box contains the currently selected mass value from the mass fragment column. When you enter a value into the ‘Extend to’ box, all possible combinations of amino acid residues that fit to this difference within the precision defined below the central table will be listed in the table. Each suggestion of residues will be listed with calculated difference, deviation from the actual difference and composition. The suggestions will be listed relative to the deviation. If you double click on a suggestion or press the ‘Transfer’ button, the selected residues will be inserted into the main fragment analyzer. Note: The residues will be inserted in the order they are shown in the ‘Extend sequence’ box, which may not correspond to the actual sequence. If you click on a mass value in the main Fragment analyzer, the value will be transferred to the ‘Start value’ box. The ‘Start’ and ‘Extend’ will also be calculated if the ‘Extend value’ is lower than the ‘Start value’. When you have a suggestion for a complete sequence, you can enter it in the ms/ms fragmentation dialog to get a list of all the normally seen fragmentation mass values (Chapter 10.1). Note: Isoleucine (Ile) is not present in the up/down mass lists as it is isobaric with leucine. When doing sequence/homology searches you have to be aware that a hit on Ile is equal to that of Leu. As the two residues have similar physical/chemical properties it is usually not of great concern.
|
|||||
|
Site last updated: February 14, 2025 |
|||||