|
Chapter A – Basic sequence handling
1 - The sequence
The basic unit of most work in GPMAW is a protein sequence. You may choose to enter a sequence manually, load it from a database or directly from the Web (e.g. see B.1/2 and C.1). However, when you work with a sequence on several occasions, you will normally save it to a file on your hard drive. The main advantage to saving sequences locally, in addition to faster access, is that the sequence saved from GPMAW may contain additional information, e.g. modified residues, cross-links, annotations etc. Note that GPMAW sequence libraries may contain multiple sequences. This works in the way that when you save to a file, which already contains one or more sequences, the new one is appended to the file, instead of replacing it.
When reading back a sequence already saved in a sequence library, there is a difference between opening a library file containing a single sequence and one containing multiple sequences. If the library file contains a single sequence, it will be read into GPMAW immediately you select the file, while you will be asked to select a sequence from a list if the library file contains multiple sequences.
Start by loading a sequence already saved in a file on the hard drive.
Select File|Open or click on the ‘Open file’ icon. This opens the ‘Open sequence library’ dialog:
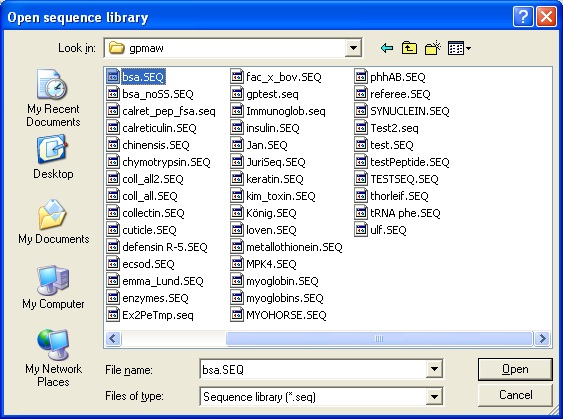
Select the ‘blood.seq’ file and you are greeted by the ‘Select sequence’ dialog.
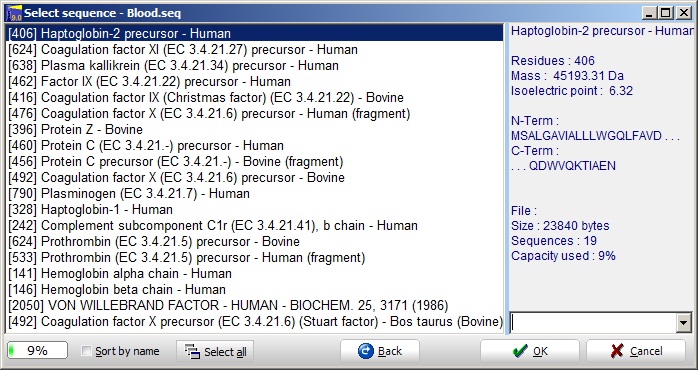
This shows a GPMAW sequence library files containing multiple sequences. As a sequence file is limited to a size of 264000 bytes (characters), there is a limit to the number of sequences that can be stored in a single library. In the present library, ‘blood.seq’, you can see (status bar at the bottom) that the file is 9% filled.
The status bar is initially green, but turns yellow and red as you get close to full capacity of the file. Furthermore, when a protein is selected, the basic information is displayed in the right-hand information box.
You may now open a sequence either by selecting it with the mouse followed by ‘OK’ (or the Enter key), or you may double-click on the sequence name to open it directly. Alternatively you can select multiple sequences by holding down the Ctrl key while selecting for a discontinuous selection. Use the Shift key for a continuous selection. You open all the selected sequence by pressing the ‘OK’ button. The ‘Back’ button returns you to the file selection dialog. The right-hand drop-down box lists the most recently opened files.
Select Protein Z, and press ‘OK’ and you will open this sequence window
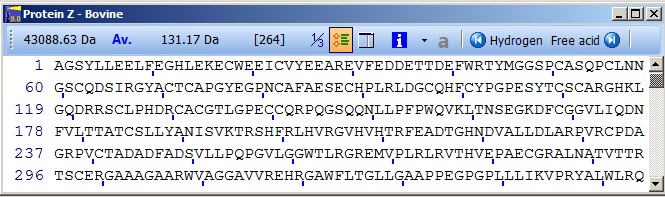
This is the basic working window of GPMAW. Most other windows will be derived from this window and are called daughter windows.
The sequence will always be displayed with the number of the first residue to the left of each line. The number of residues on a line will be the maximum number possible inside the given window, except when the ‘Multipla 5’ is turned off Setup. With the ‘Multipla 5’ turned on the number of residues on each line will be a multiple of 5 (i.e. 20, 25, 30…).
Every 10th residue is labeled with the residue number as a subscript when showing 3-letter residue code (divided by 10, i.e. residue 120 is labeled with 12) or as a small tick mark when showing 1-letter code. The feature can be turned off Setup. Use the 1/3 button in the toolbar to switch between 1- and 3- letter code.
The colors of the displayed residues can be changed for easier navigation and to indicate modifications and changes More.
2 - The toolbar

The toolbar of the sequence window contains the following:
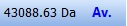 The leftmost panel shows the total mass of the protein in Daltons. The button next to it shows whether it is the average mass (Av. – blue) or monoisotopic mass (Mo. – red). You can change the mass type by clicking on the Av./Mo. button. The leftmost panel shows the total mass of the protein in Daltons. The button next to it shows whether it is the average mass (Av. – blue) or monoisotopic mass (Mo. – red). You can change the mass type by clicking on the Av./Mo. button.
The next two panels from the left can show either of two states:
 If no peptide is selected the mass of the amino acid under the cursor (i.e. the residue mass + 18 Da) will be displayed in the left-hand panel and the right-hand panel will show the residue number (if the protein is a multi-chain protein, the first chain will be labeled ‘a’, the second ‘b’ etc (i.e. 80b is residue number 80 in the second chain counting from the N-terminus). If no peptide is selected the mass of the amino acid under the cursor (i.e. the residue mass + 18 Da) will be displayed in the left-hand panel and the right-hand panel will show the residue number (if the protein is a multi-chain protein, the first chain will be labeled ‘a’, the second ‘b’ etc (i.e. 80b is residue number 80 in the second chain counting from the N-terminus).
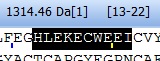 If part of the sequence is highlighted, the left panel shows the mass of the selected peptide and the right panel shows the first and last residues of the selection (see next section). If part of the sequence is highlighted, the left panel shows the mass of the selected peptide and the right panel shows the first and last residues of the selection (see next section).
Note that the mass value displayed is M, not M+H.
 The 1/3 button toggles between 1- and 3-letter code (i.e. KTA vs Lys-Thr-Ala). When you toggle between the two modes, all selections, highlights, coloring, links etc. are conserved. The default setting is done in Setup; separately on the ‘Peptide’ AND the ‘Display’ page for the peptide and sequence windows respectively. The 1/3 button toggles between 1- and 3-letter code (i.e. KTA vs Lys-Thr-Ala). When you toggle between the two modes, all selections, highlights, coloring, links etc. are conserved. The default setting is done in Setup; separately on the ‘Peptide’ AND the ‘Display’ page for the peptide and sequence windows respectively.
 Line distance. When the button is pressed, the distance between the sequence lines will be increased for easier viewing. Line distance. When the button is pressed, the distance between the sequence lines will be increased for easier viewing.
 The ‘Frames’ button toggles the display of an information frame to the left of the main sequence window. This window is dynamic, as the content of the frame will be updated when the content/selections are changed. The ‘Frames’ button toggles the display of an information frame to the left of the main sequence window. This window is dynamic, as the content of the frame will be updated when the content/selections are changed.
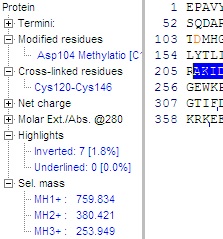 The information available is close to the same information given in the ‘sequence information window’ (see below), but in the frame it is updated dynamically. The individual terms are initially hidden, but can be expanded by clicking on the small ‘+’, changing it into a ‘-‘. The information available is close to the same information given in the ‘sequence information window’ (see below), but in the frame it is updated dynamically. The individual terms are initially hidden, but can be expanded by clicking on the small ‘+’, changing it into a ‘-‘.
Information:
Termini: Name and composition of the N- and C-termini.
Modified residues: Name, position and composition of all individually modified residues (not the ones changed globally through the mass table).
Cross-linked residues: Residues that are cross-linked, typically cysteine residues (see B-3).
Net charge: The theoretical charge of the protein at pH 2.0, 7.0 and user-selected pH (Setup).
Molar Ext./Abs.@280: Theoretical extinction coefficient / absorption of the protein at 280 nm.
Highlights: Percentage of the sequence, which is inverted (highlighted) or underlined, updated dynamically.
Sel. mass: Mass of the selected (highlighted) part of the sequence. Shown as singly, doubly and triply charged peptide (i.e. residue mass + 18 + charge). Updated dynamically. Note that unlike the peptide mass shown in the toolbar of the sequence window, this is the charged ion. More on peptide selections in part 3.
As can be seen in the sequence part of the figure, modified residues are colored (red). More
 The white on blue ‘i’ opens the sequence information window, which gives you file and statistical information, calculated indices (pI, absorption etc.), amino acid composition and multiply charged masses. The drop-down arrow open a menu giving direct access to the various pages in the information window. The white on blue ‘i’ opens the sequence information window, which gives you file and statistical information, calculated indices (pI, absorption etc.), amino acid composition and multiply charged masses. The drop-down arrow open a menu giving direct access to the various pages in the information window.
 The next button is a shortcut to the annotation page. If the annotation is empty, the ’a’ in the button will be gray, if the annotation page contains text, it will be green, blue or red depending on the content of the annotation (Swiss-Prot, Entrez or unknown format respectively). The next button is a shortcut to the annotation page. If the annotation is empty, the ’a’ in the button will be gray, if the annotation page contains text, it will be green, blue or red depending on the content of the annotation (Swiss-Prot, Entrez or unknown format respectively).
 The next two buttons show the status of the N- and C-terminal respectively (click to edit). The next two buttons show the status of the N- and C-terminal respectively (click to edit).
 The Coverage button only appears when the sequence has a corresponding coverage map associated. This can be generated from mass searches, see later. The Coverage button only appears when the sequence has a corresponding coverage map associated. This can be generated from mass searches, see later.
 The Close button closes the sequence window and all daughter windows (i.e. peptide, cleavage, search and graphical windows) The Close button closes the sequence window and all daughter windows (i.e. peptide, cleavage, search and graphical windows)
Main toolbar – Seach & cleavage section:
 Two sections of the main toolbar are of direct interest to the sequence window, the Control section (described later in Mass calculations) and the Search and cleavage section: Two sections of the main toolbar are of direct interest to the sequence window, the Control section (described later in Mass calculations) and the Search and cleavage section:
 Color residues. You can color residues in the sequence in up to three different colors. Click on the main button to enter residues or sequences (motifs) in specific colors. Alternatively select the drop-down list for quick access to the most common residues (e.g. Lys+Arg for tryptic cleavages sites; Phe, Tyr + Trp for chymotryptic cleavage sites etc.). More (Coloring residues) Color residues. You can color residues in the sequence in up to three different colors. Click on the main button to enter residues or sequences (motifs) in specific colors. Alternatively select the drop-down list for quick access to the most common residues (e.g. Lys+Arg for tryptic cleavages sites; Phe, Tyr + Trp for chymotryptic cleavage sites etc.). More (Coloring residues)
 The magnifying glass is a shortcut to peptide mass searching of the protein. More The magnifying glass is a shortcut to peptide mass searching of the protein. More
 Ms/ms search. Search the protein, list of proteins or database using peak lists in either mgf, dta or pkl format. Ms/ms search. Search the protein, list of proteins or database using peak lists in either mgf, dta or pkl format.
 The scissors are a shortcut to cutting up the protein into peptides. This is usually done using proteolytic enzymes, but may also be carried out chemically. The only requirement is that the process can be specified relative to specific residues. The small down-arrow button opens a menu enabling you to select enzyme cleavage in a single click. Note that the bottom part of the menu enables you to specify one missed cleavage, and/or to digest all opened sequence windows. More The scissors are a shortcut to cutting up the protein into peptides. This is usually done using proteolytic enzymes, but may also be carried out chemically. The only requirement is that the process can be specified relative to specific residues. The small down-arrow button opens a menu enabling you to select enzyme cleavage in a single click. Note that the bottom part of the menu enables you to specify one missed cleavage, and/or to digest all opened sequence windows. More
 Fragment button. Create (ms/ms) fragments of your protein. If part of the main sequence is highlighted, this part will be taken as the fragment peptide. If no selection has been made, the whole sequence is taken as input. If the sequence is longer than 50 residues you will be asked for confirmation before the first 400 residues are used as input. Fragment button. Create (ms/ms) fragments of your protein. If part of the main sequence is highlighted, this part will be taken as the fragment peptide. If no selection has been made, the whole sequence is taken as input. If the sequence is longer than 50 residues you will be asked for confirmation before the first 400 residues are used as input.
3 - Selections
 A useful feature is to determine the mass of a peptide that are part of the displayed protein. This is easily carried out in GPMAW by pointing the mouse at the first or last residue in the peptide, press the left mouse button and drag the mouse cursor across the sequence. A useful feature is to determine the mass of a peptide that are part of the displayed protein. This is easily carried out in GPMAW by pointing the mouse at the first or last residue in the peptide, press the left mouse button and drag the mouse cursor across the sequence.
 The mass of the selected peptide and the region covered will be shown in the toolbar. The mass of the selected peptide and the region covered will be shown in the toolbar.
The [1] displayed in the first pane indicates that only a single peptide has been highlighted. You can highlight multiple sequences by holding down the shift button while selecting additional regions. Up to three regions can be selected at a given time. The region covered will only be shown for the last selection, while the mass will be for the total..
You deselect all regions by clicking once in the sequence without holding down the Shift key.
The arrow keys can alter the most recently selected region:
The left/right arrow will change the position of the C-terminal residue of the selection one residue back or forward. Holding down the Ctrl key will similarly change the N-terminal residue of the selection. Holding down the Shift key will move the whole selection. This sound complicated, but is straightforward once you try it.
If you copy the sequence to the clipboard (Edit|Copy to clipboard or Ctrl+C), you will only copy any peptide(s) selected. If no part of the sequence is selected, the whole sequence will be copied to the clipboard in the format in which it is displayed (i.e. 1- or 3-letter code).
Note, when copying this way, only the sequence and not the name will be copied. Pressing Ctrl+F you will copy the sequence in FastA format (i.e. 1-letter code including the name of the sequence). If you want to copy the complete protein information or if you want to format the sequence (e.g. for a report), you should use the File|Export sequence menu option.
4 - Coloring residues.
One of the most efficient tools to examine and manipulate a sequence is to color the background of specific residues. This is done either through the ‘Highlight residues’ dialog (Search | Highlight residues (motifs)...), F4, the highlight button  (in the main toolbar) or by right-click and select ‘Highlight residues’ from the pop-up menu. (in the main toolbar) or by right-click and select ‘Highlight residues’ from the pop-up menu.
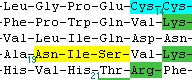 In the example (right) the basic residues have been colored one color, cysteines another color and N-glycosylation sites (N-X-S/T/C) a third color. In the example (right) the basic residues have been colored one color, cysteines another color and N-glycosylation sites (N-X-S/T/C) a third color.
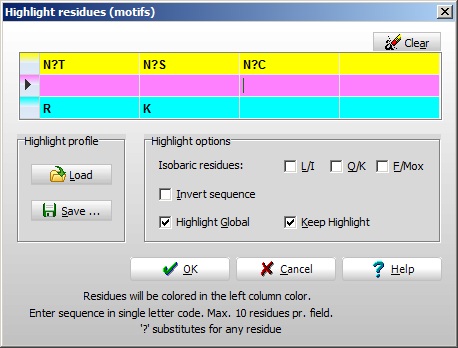
The coloring of residues is done through a simple edit box (right). Three colors are available (presented in the left-hand column) Setup, and for each color you can have four different entries (max. 10 residues in each entry). Notice that the question mark can substitute for ‘any residue’ (do not use ‘X’ as this is recognized as a specific residue – unknown). If the ‘Highlight global’ check-box is checked, all currently open sequence windows will be colored. If the ‘Keep highlight’ is not checked, the entries will be cleared whenever the ‘Highlight residues’ dialog is accessed.
‘Invert sequence’ will result in highlighting of sequences found in both N- and C-terminal directions (e.g. if entering KLGFT both the sequence KLGFT and the sequence TFGLK will be highlighted). Useful for searching for a ms/ms sequence tags (you may not know whether it is a y-ion or a b-ion series). Isobaric resides will, if checked, highlight both kinds of residues, i.e. if the ‘Q/K’ checkbox is checked and the sequence LKT is entered, the sequence LQT will also be highlighted. Again, this is for the benefit of ms/ms sequence tags.
The ‘Quickcolor’ button (the down-arrow next to the ‘Highlight’ button) opens a menu that enables you to color specific residues with a single click. The choice of selections cannot be changed by the user, but has been chosen to highlight the most common situations (e.g. highlighting R and K for a tryptic digest; C for identifying cross-links etc.).
It is also possible to highlight individual residues. This is done through the ‘Modify residue dialog’ (double-click on a residue and click on one of the colored frames at the bottom of the dialog box).
When you highlight a residue you are changing the background color. When you modify or underline residues the colors of the letters are changed (by default modified residues are colored red), so be careful when selecting colors for background as residues will ‘disappear’ if front and back colors are identical.
The colors used to highlight residues can be changed by the user Setup (Colors page).
Underlines.
Underlines are different from highlights and can be a different way of drawing attention to specific residues/sequences:
1) The can be persistent, i.e. you can save the underline information along with the sequence (File|Save w. highlights).
2) You only have a single color to work with (red by default Setup).
3) Are often used to transmit information from daughter windows (i.e. mass search) to the main sequence window.
4) As they are residue-related, they are always specific to a given sequence.
The underlines are controlled from the pop-up menu in the sequence window and the relevant daughter windows. For more information see the manual and the on-line help.
|