|
SILAC
For SILAC you want to change all residues of a given kind, e.g. lysine. You then click on the composition of lysine, click on the elemental composition editor (‘calculator’). You then change the number of carbon to 0 and number of carbon 13 (Cx) to 6. Select OK and edit any other residues that need to be changed (e.g. arginine for a typical SILAC experiment).
You then select the Save as button and save the the table as a new residue mass file (e.g. with the name SILAC). Close the dialog, close GPMAW and restart the program, and the new mass file will be available for selection in the drop-down box in the main toolbar.
AQUA
Working with both heavy and normal residues:
If you need to work with normal and heavy residues in the same sequence, you need to define the heavy residues as new residues, using the positions 21 to 30 in the residue mass table. Remember that each residue needs a unique 1-letter code to function.
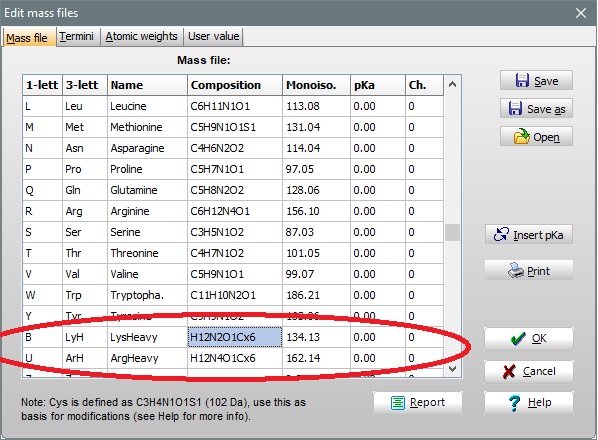
You then have to edit your sequence to put the heavy residues into place, e.g. copy the composition from a normal residue. In the example above the single letter codes B and U have been defined as the heavy lysine and arginine respectively.
Now either save the mass file using the usual name (works as normal as the heavy residue have new names) or you can use Save as to save under a different name. To use the heavy residues, just specify them using the new single residue letters, e.g. ASNLK -> ASNLB - this will expand in three letter code to Ala-Ser-Asn-Leu-Lys -> Ala-Ser-Asn-Leu-LyH.
All heavy
If you grow your recombinant protein in media with 15 as the only nitrogen source, you can exchange all nitrogens with the heavy variant. This is most easily simulated in GPMAW by clicking the 15N button on the Atomic weights page (see first picture). As this can be done ‘on-the-fly’, you do not have to save the mass file when changing.
Various
If you ‘just’ want to compare the mass values of normal and heavy residues, you can do that in the peptide list (from a digest) by using the secondary mass table feature of the peptide window (or the ‘Alternate mass file’).
|